Small Molecule Discovery Platform
- DNA-Encoded Library Technology
- DEL platform-based optimization of SIRT3 modulators

DNA-Encoded Libraries : New Chemical Entities
- Ultrahigh-throughput drug discovery using DNA-encoded library (DEL) techniques. Mixture of various molecules identified by the code of life : DNA
- Orders of magnitude larger and faster than traditional high-throughput small molecules screening: DEL HTS : 50 experiments to test 1 million compounds for 50 specific targets, compare to 50 millions experiments with classical methods
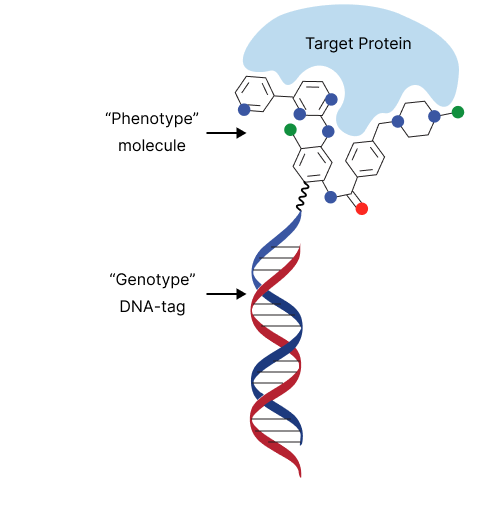
‣ chemists and biologists team dedicated to generate DNA libraries with ‘split and pool’ and/or perform selection with protein targets
‣ dedicated laboratory to perform chemistry and biology experiments as well as massively parallel NGS sequencing and data analyses
‣ Integration with computational biophysics and machine learning methods to accelerate optimization of small molecule designs compared to DEL alone
Applications
- Identification of new small molecules binding to a target biomacromolecule and exert a desired pharmaceutical effect (inhibition, activation, or protein degradation)
-
Enabled CCM Bio Rx Programs:
- Sirtuin Activators, a unique mode of upregulation of a class of enzymes that are a master regulators involved in both age-related metabolic conditions and neurodegenerative disorders – CCM-SRX
- PROTACs for Receptor Tyrosine Kinases, a recently developed, superior strategy for overcoming resistance to some of the biggest blockbuster cancer drugs through targeted protein degradation – CCM-EGX
- cGAS Inhibitors, Direct KRAS Inhibitors & Translational Readthrough Compounds discovered through virtual screening, molecular dynamics simulation, & machine learning (QSAR)
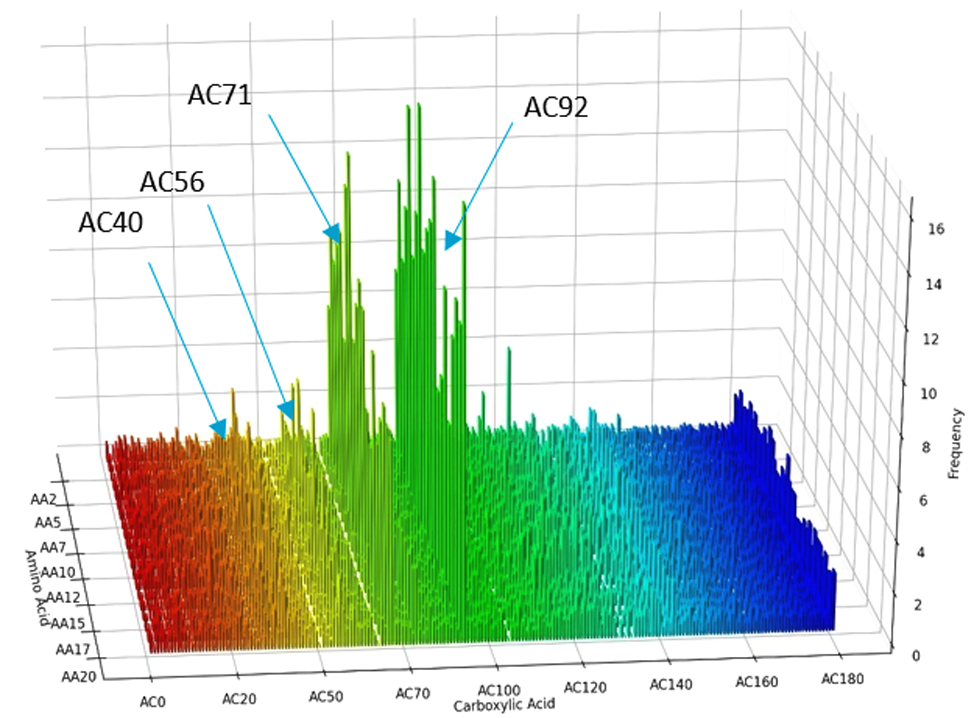
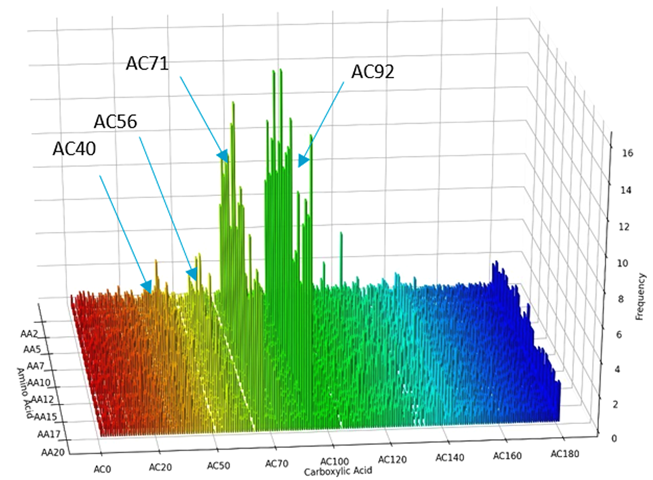
Case Study: Application to the discovery of new activators/inhibitors of Sirtuin3, a key regulator of mitochondrial metabolism.
We produced and screened a DNA encoded library consisting of the coupling of amino acids with carboxylic acids.
Patents:
- Methods for the design of mechanism-based sirtuin activating compounds.US Patent 11,118,211 B2 (2021)
- Methods for the design of mechanism-based sirtuin activating compounds. US Patent 11,459,597 (2022)
- Methods for the design of nonallosteric sirtuin activating compounds. US patent application 2022/0002778 A1 (2021)
- Sirtuin modulating compounds and applications thereof.
US patent application 18/090,953 (2022)
- Discovery of novel compounds as potent activators of Sirt3. Célina Reverdy, Gaetan Gitton, Xiangying Guan, Indranil Adhya, Rama Krishna Dumpati, Samir Roy, Santu Chall, Anisha Ghosh, Gauthier Errasti, Thomas Delacroix and Raj Chakrabarti. Bioorganic and Medicinal Chemistry 73 116999 (2022)
Drug Delivery Platform
- Drug Delivery Technology
- Polyaminoacid and Lipid Nanoparticles for Passive Drug Delivery
- Polyaminoacid and Lipid Nanoparticles for Active Drug Delivery

New Excipients
- New polymers (PolyAminoacids - PAA) research and development
- Excipients with excellent biodegradability, easy functionalization, easy conjugation, bioavailability properties.
- Polymers characterization, process development and scale-up
- Unique know-how and integration from raw materials to PolyAminoAcids production

New Drug Delivery Systems
- Drug delivery systems with versatile architecture and functionality
- Superior biodegradability and biocompatibility
- High cargo loadings and improved bioavailability of your APIs
- Modulation of interactions with biological barriers allowing to explore multiple administration routes
Enabled CCM Bio Programs
- Nanodelivery of cytotoxic APIs (chemotherapy) constituting the next-generation solution to targeted chemotherapy
- Nanodelivery of gene therapy APIs including KRAS mutant-selective RNA interference – CCM-MRX
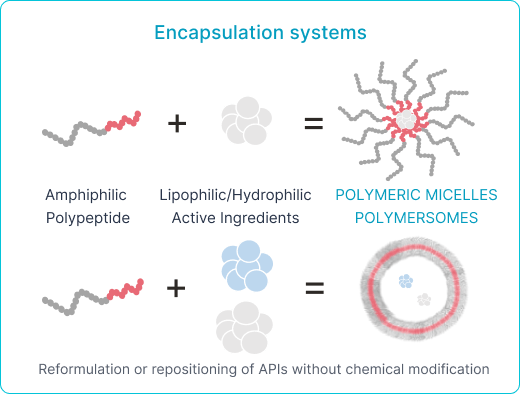
Polyaminoacids for PASSIVE drug delivery

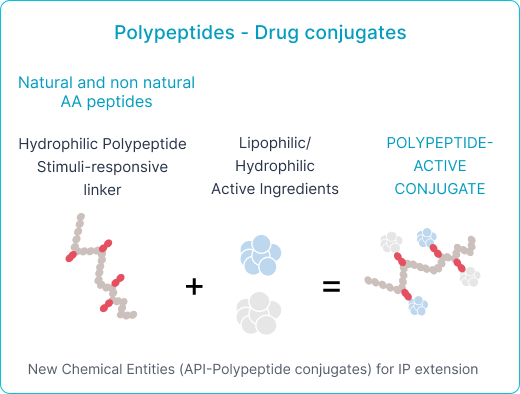
Patents:
- Amphiphilic polyamino acid block copolymers and nanoparticles thereof for drug delivery applications. EP patent application 22305766.2 – 1102, Filed 2022
- Improving aqueous solubility of paclitaxel with polysarcosine-b-poly(γ-benzyl glutamate) nanoparticles. Coralie Lebleu, Laetitia Plet, Florène Moussy, Gaëtan Gitton, Rudy Da Costa Moreira, Barbara Burlot, Rodolphe Godiveau, Aïnhoa Merry, Sébastien Lecommandoux, Gauthier Errasti, Christiane Philippe, Thomas Delacroix and Raj Chakrabarti. International Journal of Pharmaceutics 631 122501 (2023)
Polyaminoacids for ACTIVE drug delivery
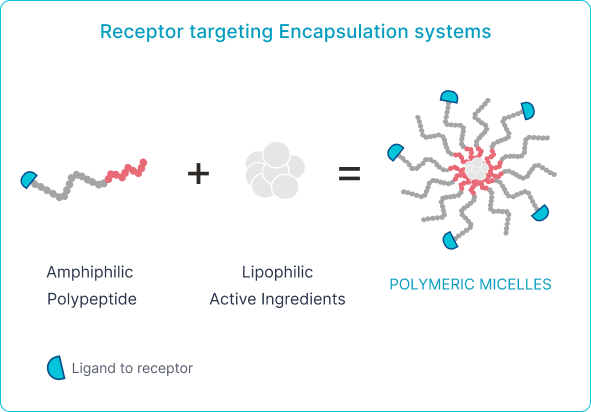

Engineered peptide or antibody (identified by Phage Display) conjugated to PAA micelle
Biological interaction between ligands on the surface of nanoparticles and the cell target Several types of ligands could be used for facilitating active targeting of NPs : bind to specific receptors on the surface of the target cells and increase cellular uptake of drug-containing NPs and increase therapeutic efficacy.
Additional capabilities:
Delivery of gene therapy APIs (nucleic acids -- DNA/RNA drugs) by cationic polymers through conjugation or encapsulation
Patents:
- New EGFRvIII-binding peptides, conjugates and uses thereof. European patent application B77140EP; International patent application PCT/EP2022/087500, Filed 2022
- Identification of a novel peptide ligand for the cancer-specific receptor mutation EGFRvIII using high-throughput sequencing of phage-selected peptides. Sourour Mansour, Indranil Adhya, Coralie Lebleu, Rama Dumpati, Ahmed Rehan, Santu Chall, Jingqi Dai, Gauthier Errasti, Thomas Delacroix and Raj Chakrabarti. Scientific Reports 12 20725 (2022)
Biologics Discovery Platform
- Machine Learning-enabled Phage Display Technology
- CASE STUDY (active Drug Delivery Application): EGFR vIII peptide binder

Phage Display: New Biologics
- Ultrahigh-throughput drug discovery using Phage Display: a combinatorial biology technique
- Identify hit peptides and antibodies as therapeutic agents, vaccines, diagnostic reagents, as well as gene and drug delivery systems
- Ability to generate very high diverse exogenous peptides or proteins displayed on the surface of the phage using standard molecular biology
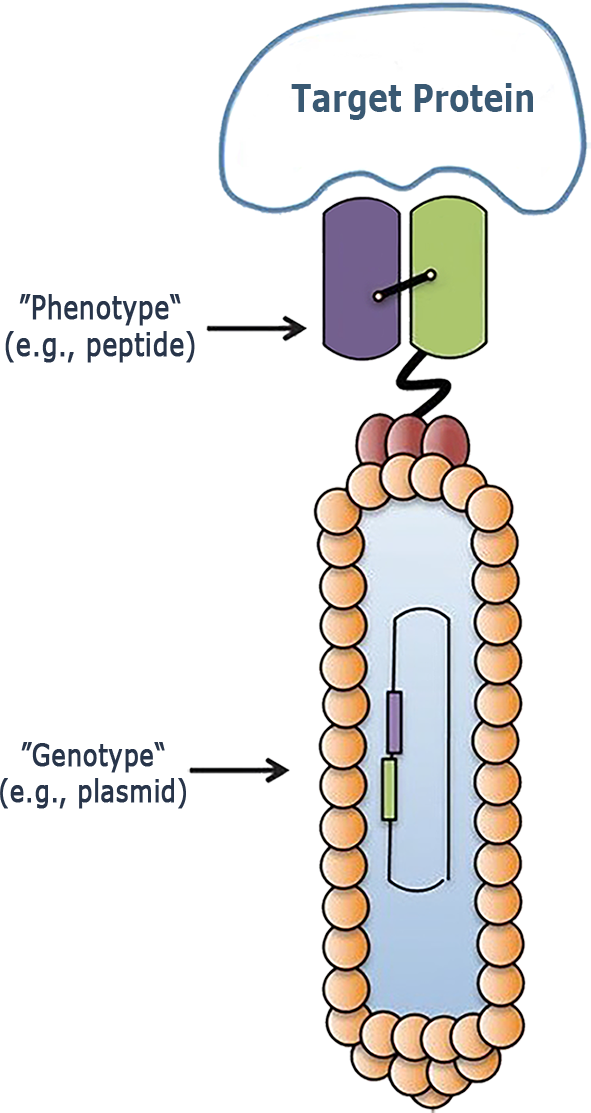
‣ A biologists team dedicated to generate and screen P.D. libraries
‣ A dedicated laboratory to perform chemistry and biology experiments as well as massively parallel NGS sequencing and data analyses
‣ Integration with computational biophysics and machine learning methods to accelerate optimization of small molecule designs compared to DEL alone
Applications
- Identification of new molecules binding to a target biomacromolecule and exert a desired pharmaceutical effect
-
Enabled CCM Bio Rx Programs:
- EGFR-targeted active drug delivery for anticancer chemotherapy APIs including paclitaxel and SN-38, through targeting of cancer-specific epitopes such as EGFRvIII in glioblastoma (brain cancer)
- KRAS mutant-selective gene therapy using nanocarrier delivery vehicles actively targeted toward cancer cells -- CCM-MRX
CASE STUDY (active drug delivery application) : EGFR vIII peptide binder
Background:
One of the few known cancer-specific cell surface markers is the epidermal growth factor (EGFR) tyrosine kinase receptor mutation variant III (EGFRvIII). Tumor-targeting ligands such as peptides and antibodies may effectively aid certain cytotoxic agents (either biological or synthetic) to deliver to the tumor cells, thereby improving therapeutic efficacy while limiting the exposure of normal tissues to the cytotoxic agents
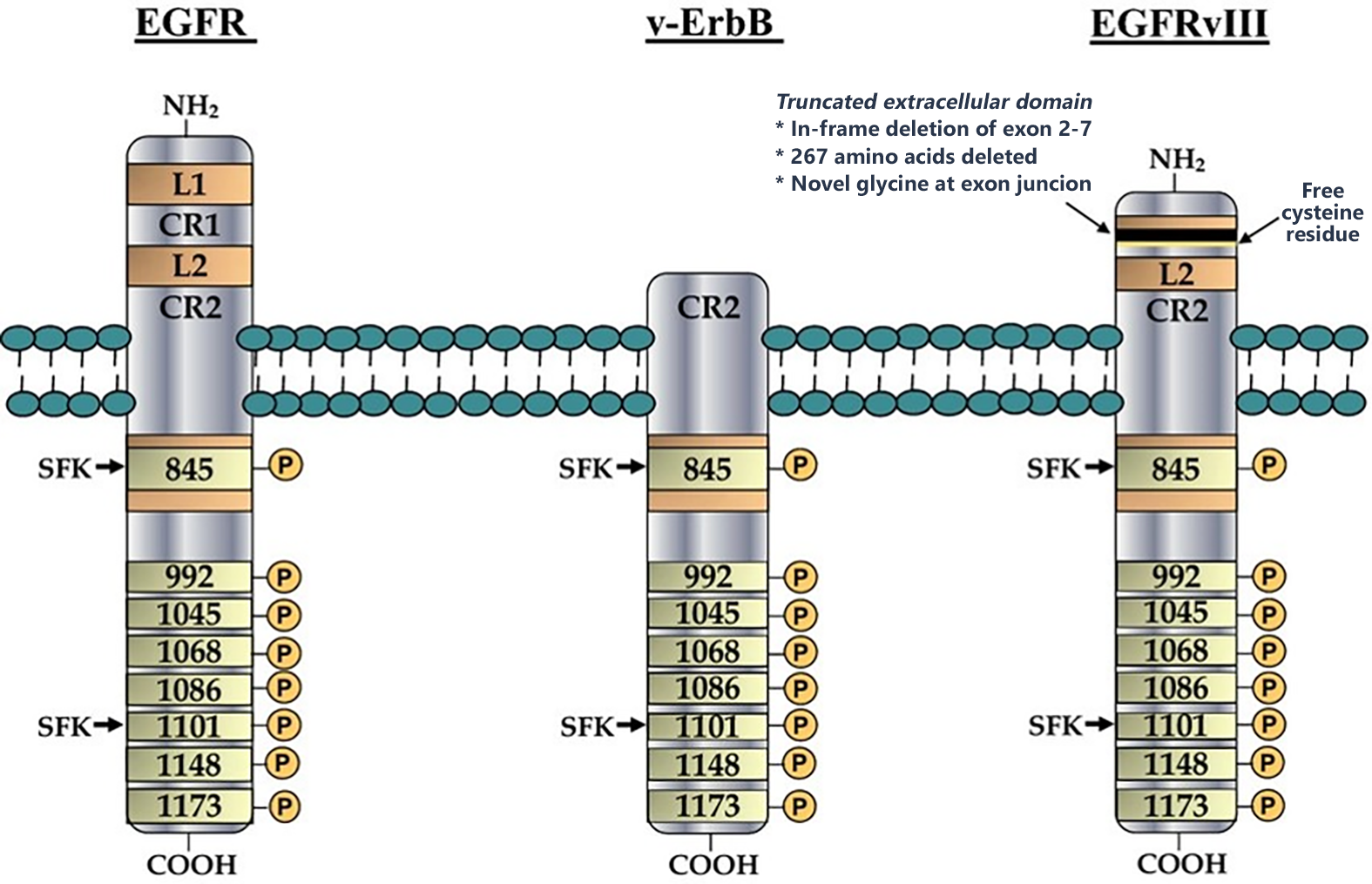
Project goal: identification of novel peptide ligands targeted EGFRvIII, which could be used as a targeting ligand assisting specific delivery of cytotoxic drug specifically into the tumor vasculature, tumor microenvironment or into the cancer cells
Patents:
- New EGFRvIII-binding peptides, conjugates and uses thereof. European patent application B77140EP; International patent application PCT/EP2022/087500, Filed 2022
- Identification of a novel peptide ligand for the cancer-specific receptor mutation EGFRvIII using high-throughput sequencing of phage-selected peptides. Sourour Mansour, Indranil Adhya, Coralie Lebleu, Rama Dumpati, Ahmed Rehan, Santu Chall, Jingqi Dai, Gauthier Errasti, Thomas Delacroix and Raj Chakrabarti. Scientific Reports 12 20725 (2022)
Enzyme Engineering Platform
- Ultrahigh-throughput Droplet Microfluidics
- Droplet Microfluidics: Further Details

Protein/Enzyme Engineering: New Enzymes
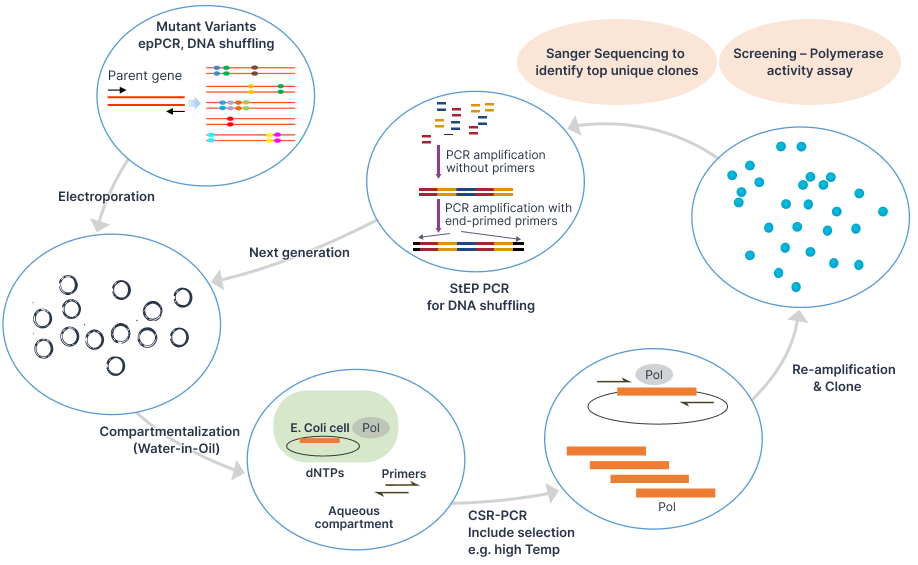
- Enzyme engineering using ultrahigh-throughput droplet microfluidics
- Upstream processing: Proteins/biologics production by fermentation; Nucleic acids production by biocatalysis
- Downstream processing: Purification, scale-up and characterization
- Antibodies, bioconjugates manufacturing
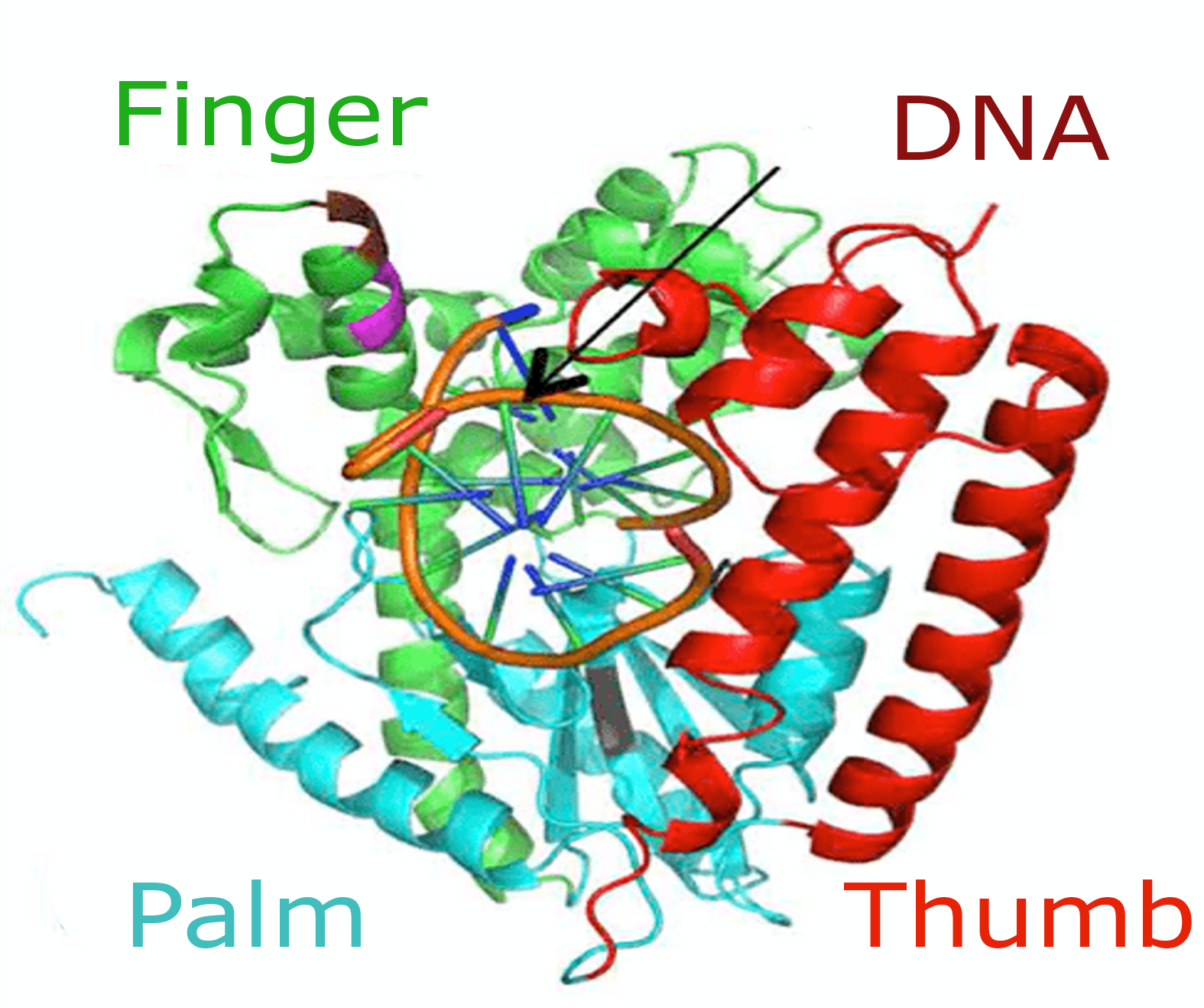
‣ A biologists team dedicated to produce, purify and characterize proteins
‣ A dedicated laboratory to perform molecular biology and enzyme engineering, production and characterization
‣ Integration with computational biophysics and machine learning methods to accelerate optimization of enzyme designs beyond directed evolution alone
Applications
- Production and characterization of proprietary polymerase enzymes, used in PCR technology, DNA sequencing and enzymatic DNA/RNA (gene therapy) manufacturing
-
Enabled CCM Bio Programs:
- Companion Cancer Dx tests (liquid biopsy) for CCM Bio’s Rx programs, using droplet digital PCR (ddPCR)
- Companion Dx tests (liquid biopsy) for CCM Bio’s Rx programs, using next-generation DNA sequencing (NGS), for both Cancer and Prenatal Applications (IVF)
- Enzymatic DNA synthesis and manufacturing, for efficient manufacturing of gene therapy APIs
Droplet Microfluidics: Further Details
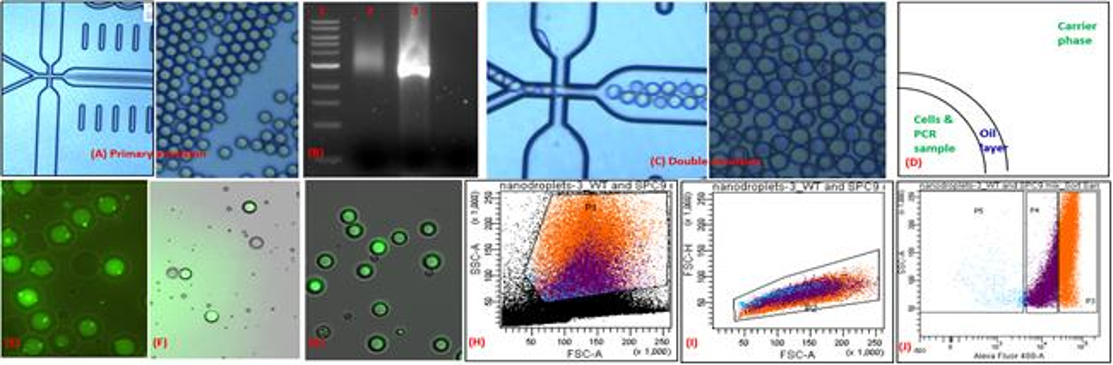
- System and methods for determination of temperature cycling protocols for polymerase chain reactions. US Patent 10,861,586 B2 (2020)
- System and methods for determination of temperature cycling protocols for polymerase chain reactions. EP Patent 3,000,366 B1 (2020)
- System and methods for determination of temperature cycling protocols for polymerase chain reactions: optimally controlled PCR. US patent application 2019/0182944 A1, Filed 2019
- System for determination of temperature cycling protocols for polymerase chain reactions: polymerase extension. US patent application 2019/0279739 A1, Filed 2019
- System for determination of temperature cycling protocols for polymerase chain reactions: oligonucleotide annealing. US patent application 2019/0270189245, Filed 2019
- System for determination of temperature cycling protocols for polymerase chain reactions: DNA melting. US patent application 2019/0295686 A1, Filed 2019
- Polymerases for mixed aqueous-organic media and uses thereof, International patent application PCT/US2022/011076. Filed 2022
- Compositions and methods for enhancing PCR and other polynucleotide amplification reactions. US Patent 6,949,368 (2005)
- Compositions and methods for enhancing polynucleotide amplification reactions. US Patent 7,276,357 (2007)
- Chemical PCR: Compositions and methods for enhancing polynucleotide amplification reactions. US Patent 7,772,383 (2010)
- DNA amplification in mixed aqueous - organic media: chemical analysis of leading polymerase chain reaction compositions. Jalel Neffati, Ioanna Petrounia, Rudy D. Moreira and Raj Chakrabarti. bioRxiv 2021.10.02.462877 (2021)
- Evolution of Organic Solvent-Resistant DNA Polymerases. Mohammed Elias, Xiangying Guan, Devin Hudson, Rahul Bose, Joon Kwak, Ioanna Petrounia, Kenza Touah, Sourour Mansour, Peng Yue, Gauthier Errasti, Thomas Delacroix, Anisha Ghosh, and Raj Chakrabarti. ACS Synthetic Biology 10.1021/acssynbio.2c00515 (2023)
- Applying ultrahigh-throughput screening and massively parallel data analysis to identify improved polymerase variants in the presence of organic solvents. Xiangying Guan, Devin Hudson, Rahul Bose, M Elias, J Kwak, Ioanna Petrounia, K Touah, S Mansour, Gauthier Errasti, Thomas Delacroix, Anisha Ghosh and Raj Chakrabarti. (2022)
- Sequence-dependent theory of oligonucleotide hybridization kinetics. Karthikeyan Marimuthu and Raj Chakrabarti. J Chem Phys 140 (17) 175104 (2014)
- Dynamics and control of DNA sequence amplification. Karthikeyan Marimuthu and Raj Chakrabarti. J Chem Phys 141 (16) 164119 (2014)
- Sequence-dependent biophysical modeling of DNA amplification. Karthikeyan Marimuthu, Chaoran Jing and Raj Chakrabarti. Biophys J 107 (7) 1731-1743 (2014)
- The enhancement of PCR amplification by low molecular weight amides. Raj Chakrabarti and CE Schutt. Nucleic Acids Res. 29 (11) 2377-2381 (2001)
- The enhancement of PCR amplification by low molecular weight sulfones. Raj Chakrabarti and CE Schutt. Gene 274 (1-2) 293-298 (2001)
- "Novel PCR-enhancing compounds and their modes of action," R. Chakrabarti In: PCR Technology: Current Innovations, 2nd Edition. Ed. T. Weissensteiner, H.G. Griffin and A. Griffin, CRC Press: Boca Raton, FL (2003) [invited article]


